PROPT Sequential Activation of Metabolic Pathways
From TomWiki
Jump to navigationJump to search
|
This page is part of the PROPT Manual. See PROPT Manual. |
a Dynamic Optimization Approach 2009, Diego A. Oyarzuna, Brian P. Ingalls, Richard H. Middleton, Dimitrios Kalamatianosa
Problem description
The problem is described in the paper referenced above.
% Copyright (c) 2010-2010 by Tomlab Optimization Inc.
% Unbranched Pathway (n=4) with MM kinetics
kcat0 = 1; kcat1 = 2; kcat2 = 4; kcat3= 3;
Km = 1; s0 = 5; V = 0.2;
Et = 1; Umax = 1; lam = 0.5; tfmax = 50;
% Terminal constraints, compatible with steady state flux V
s1f = 0.65; s2f = 0.32; s3f = 0.27;
e0f = V*(Km/kcat0/s0 + 1/kcat0);
e1f = V*(Km/kcat1/s1f + 1/kcat1);
e2f = V*(Km/kcat2/s2f + 1/kcat2);
e3f = V*(Km/kcat3/s3f + 1/kcat3);Problem setup
N = [30 128]; % Number of collocation points
toms t t_f
warning('off', 'tomSym:x0OutOfBounds');
for n = N
p = tomPhase('p', t, 0, t_f, n);
setPhase(p);
tomStates s1 s2 s3 e0 e1 e2 e3
tomControls u0 u1 u2 u3
% Initial guess
if n == N(1)
x0 = {t_f == 2
icollocate({
[s1;s2;s3] == 0
e0 == t/t_f
e1 == (t-0.2)/t_f
e2 == (t-1)/t_f
e3 == (t-1.2)/t_f
})
collocate({u0 == 1
[u1;u2;u3] == 0})};
else
x0 = {t_f == tf_init
icollocate({
s1 == s1_init; s2 == s2_init; s3 == s3_init
e0 == e0_init; e1 == e1_init; e2 == e2_init
e3 == e3_init})
collocate({u0 == u0_init
u1 == u1_init; u2 == u2_init; u3 == u3_init})};
end
% Box constraints
cbox = {0.1 <= t_f <= tfmax
0 <= icollocate([s1;s2;s3]) <= 100
0 <= icollocate([e0;e1;e2;e3]) <= 1
0 <= collocate([u0;u1;u2;u3]) <= Umax};
% Boundary constraints
cbnd = {initial({[s1;s2;s3] == 0; [e0;e1;e2;e3] == 0})
final({s1 == s1f; s2 == s2f; s3 == s3f;
e0 == e0f; e1 == e1f; e2 == e2f; e3 == e3f})};
% Michaelis-Mentes ODEs and path constraints
ceq = collocate({
dot(s1) == kcat0*s0*e0/(Km + s0) - kcat1*s1*e1/(Km+s1)
dot(s2) == kcat1*s1*e1/(Km+s1) - kcat2*s2*e2/(Km+s2)
dot(s3) == kcat2*s2*e2/(Km+s2) - kcat3*s3*e3/(Km+s3)
dot(e0) == u0 - lam*e0
dot(e1) == u1 - lam*e1
dot(e2) == u2 - lam*e2
dot(e3) == u3 - lam*e3});
% Objective
objective = integrate(1 + e0 + e1 + e2 + e3);Solve the problem
options = struct;
options.name = 'Metabolic Pathways, Unbranched n=4';
solution = ezsolve(objective, {cbox, cbnd, ceq}, x0, options);
% Collect solution as initial guess
s1_init = subs(s1,solution);
s2_init = subs(s2,solution);
s3_init = subs(s3,solution);
e0_init = subs(e0,solution);
e1_init = subs(e1,solution);
e2_init = subs(e2,solution);
e3_init = subs(e3,solution);
tf_init = subs(t_f,solution);
u0_init = subs(u0,solution);
u1_init = subs(u1,solution);
u2_init = subs(u2,solution);
u3_init = subs(u3,solution);Problem type appears to be: qpcon
Time for symbolic processing: 0.50311 seconds
Starting numeric solver
===== * * * =================================================================== * * *
TOMLAB - TOMLAB Development license 999007. Valid to 2011-12-31
=====================================================================================
Problem: --- 1: Metabolic Pathways, Unbranched n=4 f_k 6.092698010914424400
sum(|constr|) 0.000001117096751345
f(x_k) + sum(|constr|) 6.092699128011175500
f(x_0) 4.220507512582260600
Solver: snopt. EXIT=0. INFORM=1.
SNOPT 7.2-5 NLP code
Optimality conditions satisfied
FuncEv 1 ConstrEv 34 ConJacEv 34 Iter 22 MinorIter 1809
CPU time: 0.249602 sec. Elapsed time: 0.246000 sec.
Problem type appears to be: qpcon
Time for symbolic processing: 0.51985 seconds
Starting numeric solver
===== * * * =================================================================== * * *
TOMLAB - TOMLAB Development license 999007. Valid to 2011-12-31
=====================================================================================
Problem: --- 1: Metabolic Pathways, Unbranched n=4 f_k 6.085709050712100800
sum(|constr|) 0.000004797534668342
f(x_k) + sum(|constr|) 6.085713848246769000
f(x_0) 6.092720075212342000
Solver: snopt. EXIT=0. INFORM=1.
SNOPT 7.2-5 NLP code
Optimality conditions satisfied
FuncEv 1 ConstrEv 8 ConJacEv 8 Iter 6 MinorIter 2215
CPU time: 6.224440 sec. Elapsed time: 3.642000 sec.
end
% Collect data
t = subs(collocate(t),solution);
s1 = subs(collocate(s1),solution);
s2 = subs(collocate(s2),solution);
s3 = subs(collocate(s3),solution);
e0 = subs(collocate(e0),solution);
e1 = subs(collocate(e1),solution);
e2 = subs(collocate(e2),solution);
e3 = subs(collocate(e3),solution);
u0 = subs(collocate(u0),solution);
u1 = subs(collocate(u1),solution);
u2 = subs(collocate(u2),solution);
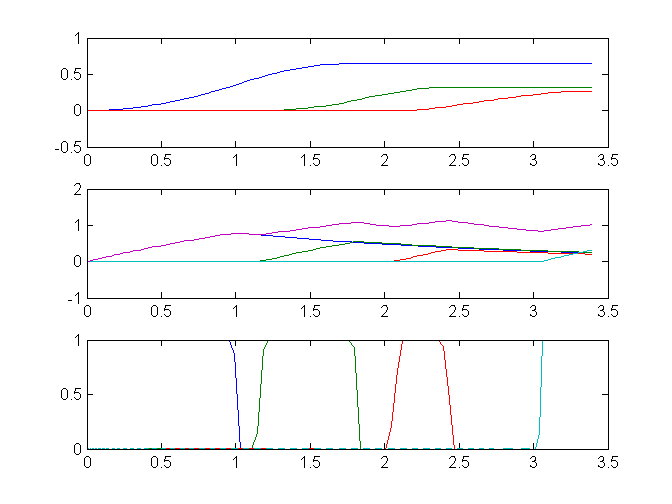
u3 = subs(collocate(u3),solution);Plot result
s = [s1 s2 s3];
e = [e0 e1 e2 e3];
r = [u0 u1 u2 u3];
subplot(3,1,1);
plot(t,[s1 s2 s3]);
subplot(3,1,2);
plot(t,[e0 e1 e2 e3 e0+e1+e2+e3]);
subplot(3,1,3);
plot(t,[u0 u1 u2 u3]);